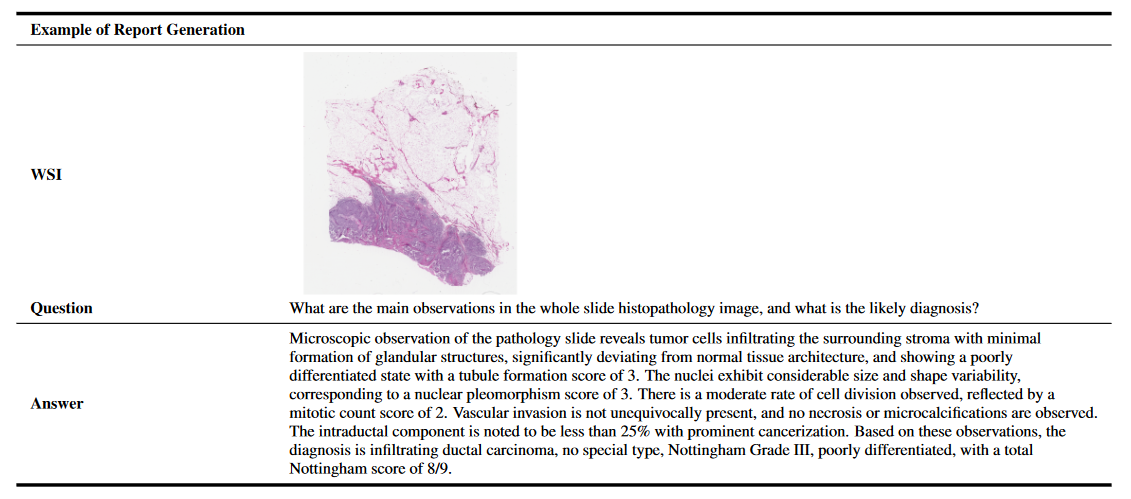
Examples of 11 Pathology VQA Tasks
Key Contributions
• We introduce WSI-Bench, the first large-scale morphology-aware benchmark for gigapixel WSI understanding and evaluation, encompassing 180k VQA pairs from 9,850 WSIs across 30 cancer types. This benchmark uniquely emphasizes morphological observations in evaluating WSI-level MLLMs.
• We propose WSI-LLaVA, a novel framework for gigapixel WSI analysis that bridges the cross-modal gap between WSIs and textual descriptions. The framework introduces a three-stage training approach: WSI-text alignment, feature space alignment, and task-specific instruction tuning.
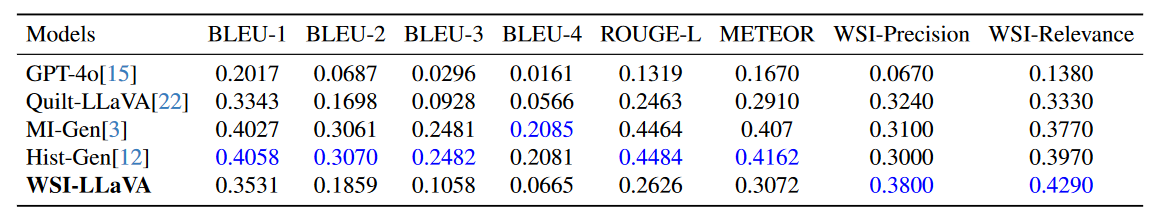
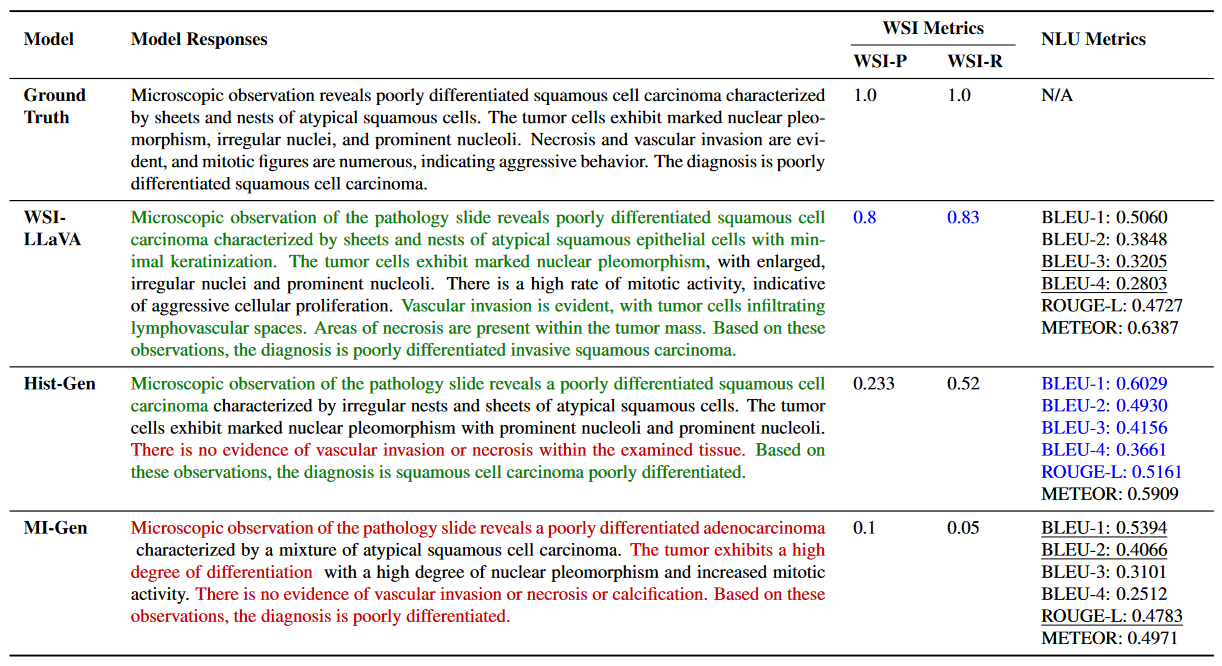
• We develop WSI-specific evaluation metrics (WSI-Precision and WSI-Relevance) that provide a more accurate assessment of model performance in pathological contexts, addressing the limitations of traditional NLU metrics by verifying claim accuracy and response relevance.
• Through comprehensive experiments, we demonstrate WSI-LLaVA’s superior performance compared to existing models, establishing a clear correlation between morphological capabilities and diagnostic accuracy.
WSI-Bench
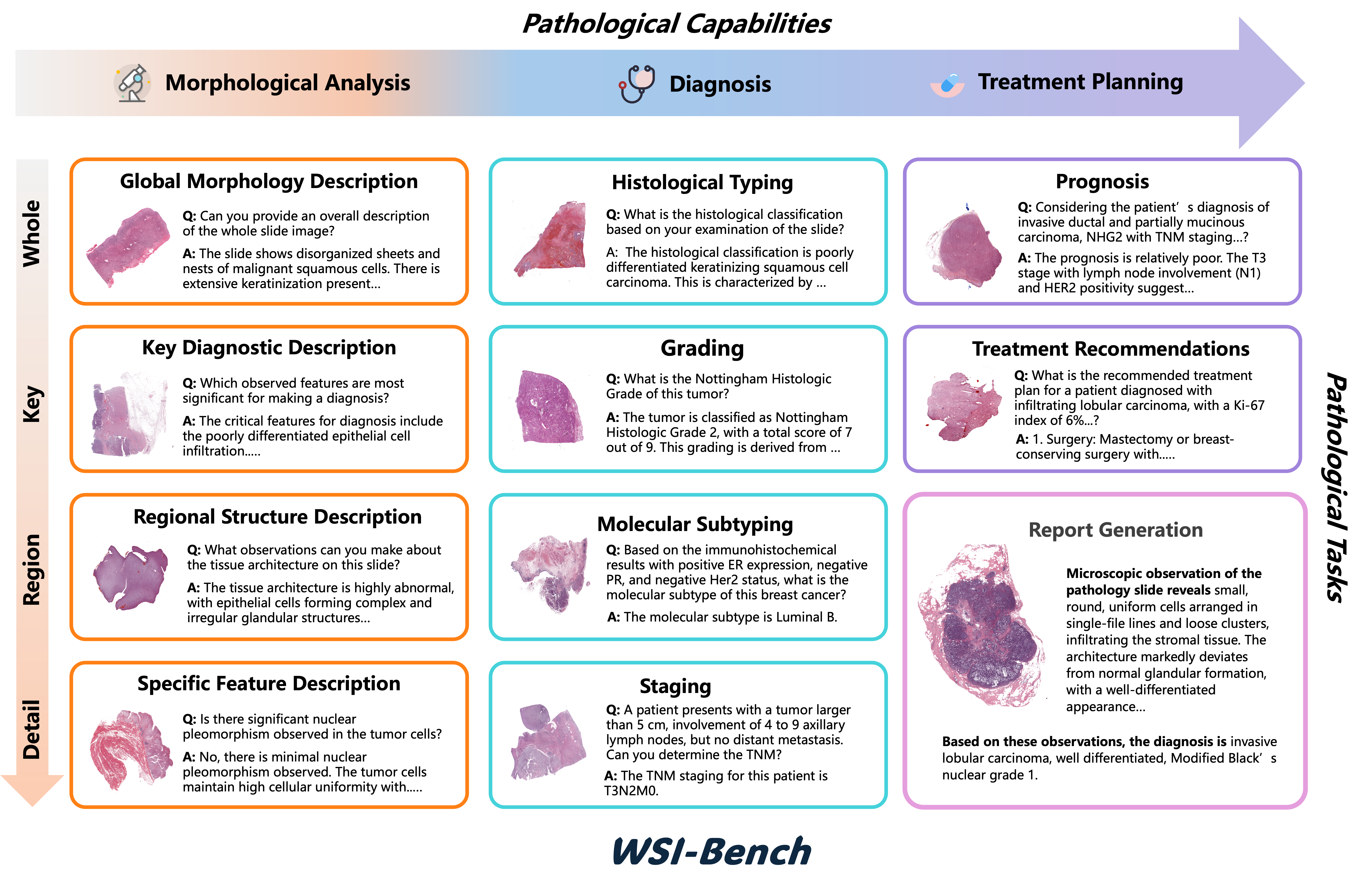
In clinical practice, pathologists rely heavily on morphological features, particularly tissue and cellular structural abnormalities, for diagnosis. Current WSI models overlook these critical details, impacting diagnostic accuracy.So,We introduce WSI-Bench, a morphology-aware benchmark for gigapixel WSI evaluation across 3 pathological capabilities and 11 tasks, which encompasses about 180k VQA pairs from 9,850 WSI across 30 cancer types, sourced from 8,368 patients.
WSI-Bench Construction
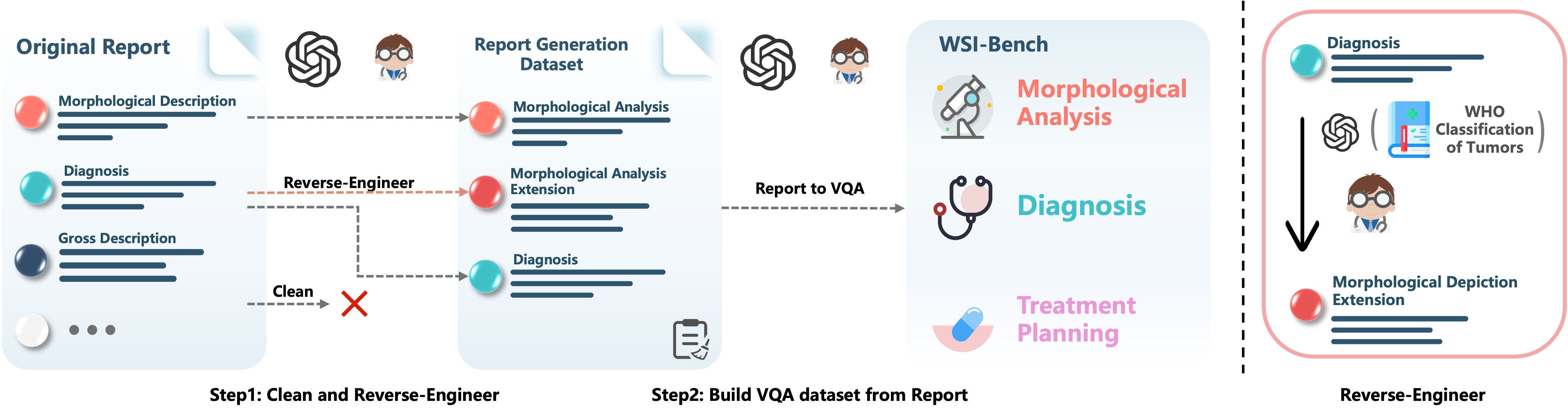
Step 1: Remove gross descriptions and IHC results from pathology reports, retaining morphological descriptions and diagnostic conclusions, then generate enriched descriptions through reverse engineering.
Step 2: Construct VQA pairs from the refined reports to support comprehensive pathological tasks, enabling diverse analytical capabilities.
Architecture
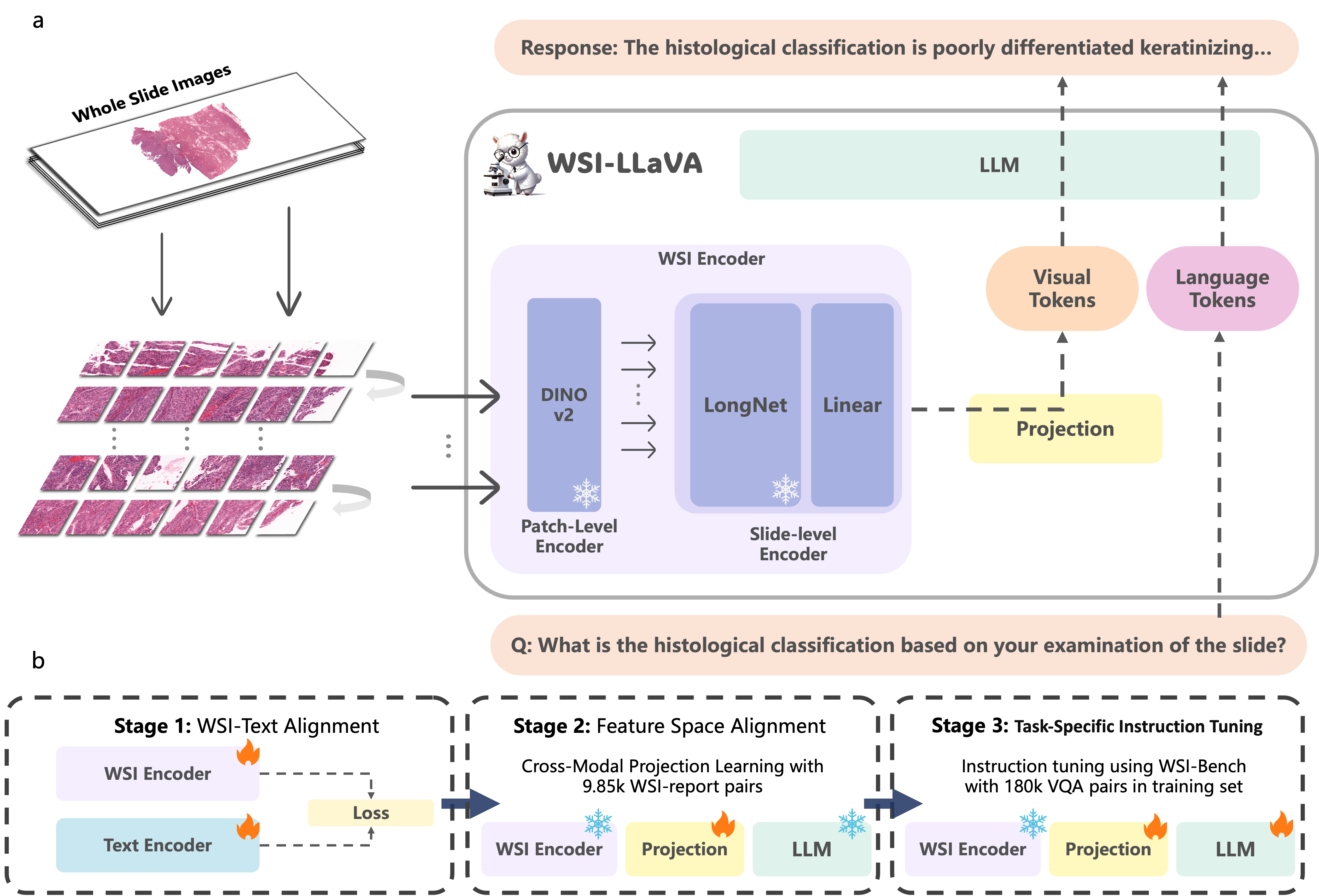
Model Architecture:The framework comprises three key components: the WSI Encoder, a projection layer, a large language model.
Training Strategy: WSI-LLaVA adopts an innovative three-stage training approach for gigapixel WSI analysis, which includes:WSI-Text Alignmen,Feature Space Alignment,Task-Specific Instruction Tuning.
WSI-Metrics
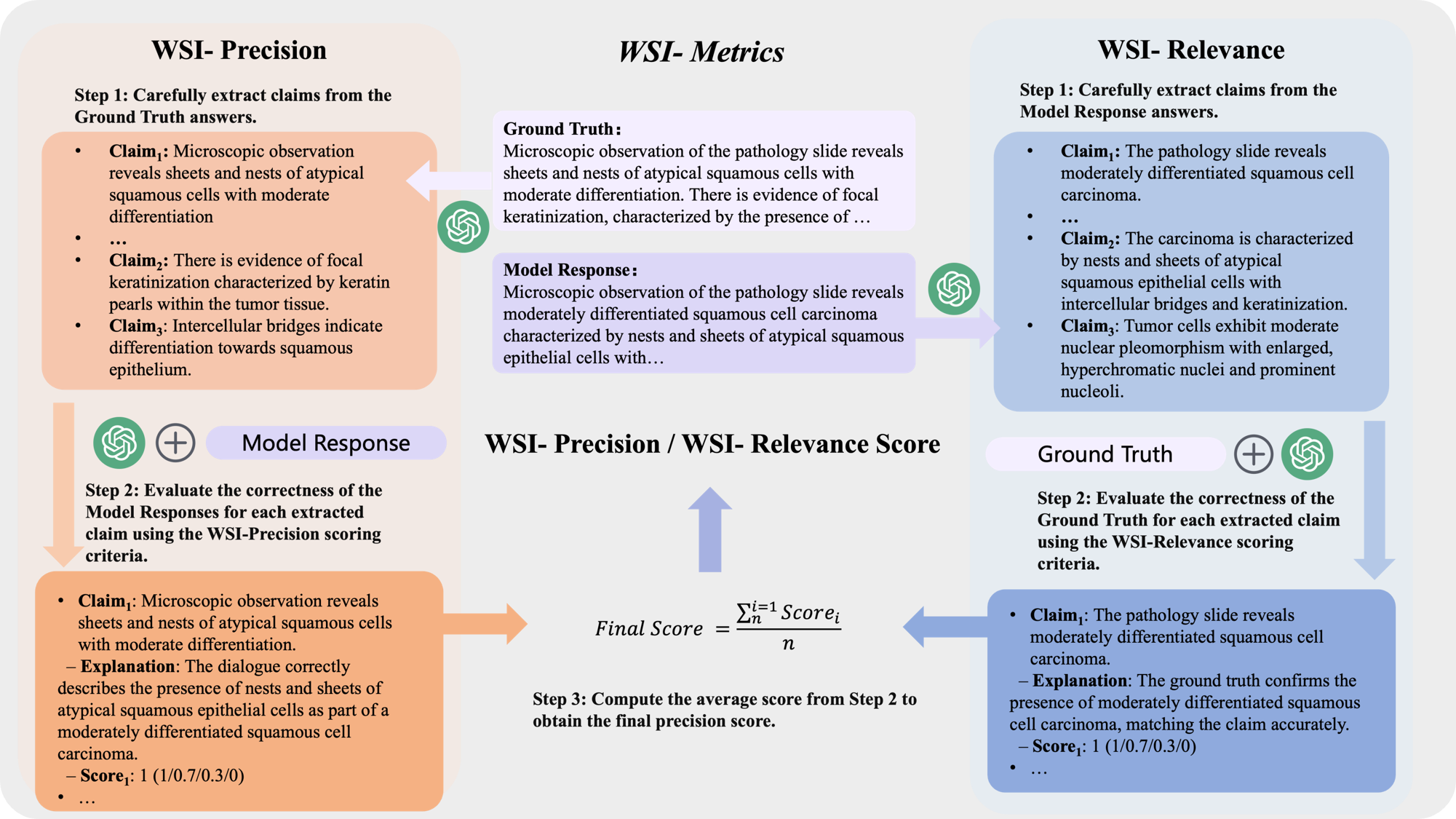
While Natural Language Understanding (NLU) metrics are commonly used to evaluate medical language tasks, they fall short in accurately assessing performance due to pathology’s complex and often similar terminology. To address this limitation, we introduce two specialized WSI metrics: WSI-Precision, which verifies the accuracy of each claim derived from the ground truth against the model’s answers. WSI-Relevance, which assesses the alignment of each claim in the model’s responses with the ground truth to ensure their relevance.
Experiments
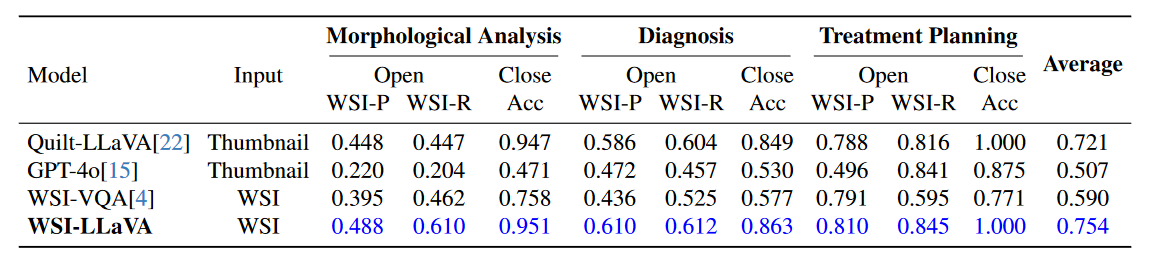
We collect various WSI-level MLLMs to evaluate on our WSI-Bench dataset. These include pecialized models for WSI report generation, such as MI-Gen and Hist-Gen, as well as models designed for pathological VQA tasks, like Quilt-LLaVA and WSI-VQA. Additionally, we assess GPT-4o’s performance to evaluate a general-purpose MLLM. For models with input size constraints (e.g., QuiltLLaVA and GPT-4o), we resize the WSIs to 1024 × 1024 pixels to fit within their input processing capabilities. To ensure a fair comparison, all WSI MLLMs are trained on WSI-Bench’s training set and evaluated on its test set.(WSI-P: WSI-Precision, WSI-R: WSI-Relevance, Acc: accuracy, open: open-ended question, and close: close-ended question.)
Citation
@article{liang2024wsi,
title={WSI-LLaVA: A Multimodal Large Language Model for Whole Slide Image},
author={Liang, Yuci and Lyu, Xinheng and Ding, Meidan and Chen, Wenting and Zhang, Jipeng and Ren, Yuexiang and He, Xiangjian and Wu, Song and Yang, Sen and Wang, Xiyue and others},
journal={arXiv preprint arXiv:2412.02141},
year={2024}
}